 |
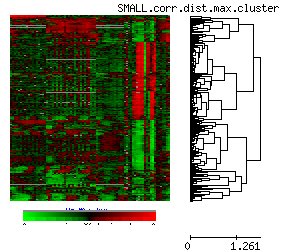
Microarray time series classification
We are utilizing kernel methods for classsification of microarray time series data. This classification of gene expression time series has many further potential applications in medicine and pharmacogenomics, such as disease diagnosis, drug response prediction or disease outcome prognosis, contributing to individualized medical treatment. |
 |
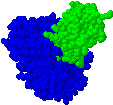
Protein function prediction
We have designed graph representations of proteins integrating sequence, structure and bio-chemical information.
We have applied graph kernels for protein function prediction on these models. Future work will aim at designing faster and more expressive graph kernels and at exploring new approaches to protein function prediction. |
 |
Subspace Clustering
Finding clusters in high-dimensional data is usually futile. But high-dimensional
data may be clustered differently in varying subspaces of the feature
space. Subspace clustering aims at finding all subspaces of high-dimensional
data in which clusters exist. |
|
RIS
A method for finding all subspaces of high-dimensional data containing
density-based clusters. |
 |
Retrieval of Feature Graphs and Activity Maps
Potential docking sites that are represented by feature graphs and activity
maps are retrieved from a 3D protein database in order to provide an efficient
filter step for the one-to-many protein docking prediction. |
|
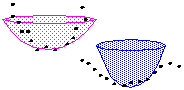
3D Shape Similarity Search in Biomolecular Databases
From a 3D protein database, molecules that have a similar 3D shape are
retrieved by using a similarity model based on 3D shape histograms. |
 |
Similarity Search for 3D Surface Segments
As a part of protein-protein docking prediction, we perform a similarity
search on 3D surface segments. Parametric surface functions including paraboloids
and trigonometric polynomials are used to approximate the surface segments. |
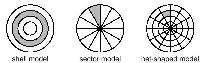 |
Histogram-Based Shape Similarity
Sector, Shell and Web Histogram Model |
 |
k-Nearest Neighbor Classification
Whereas performance is a serious problem for many k-nn classifiers,
our query processor efficiently supports this data mining technique. |

